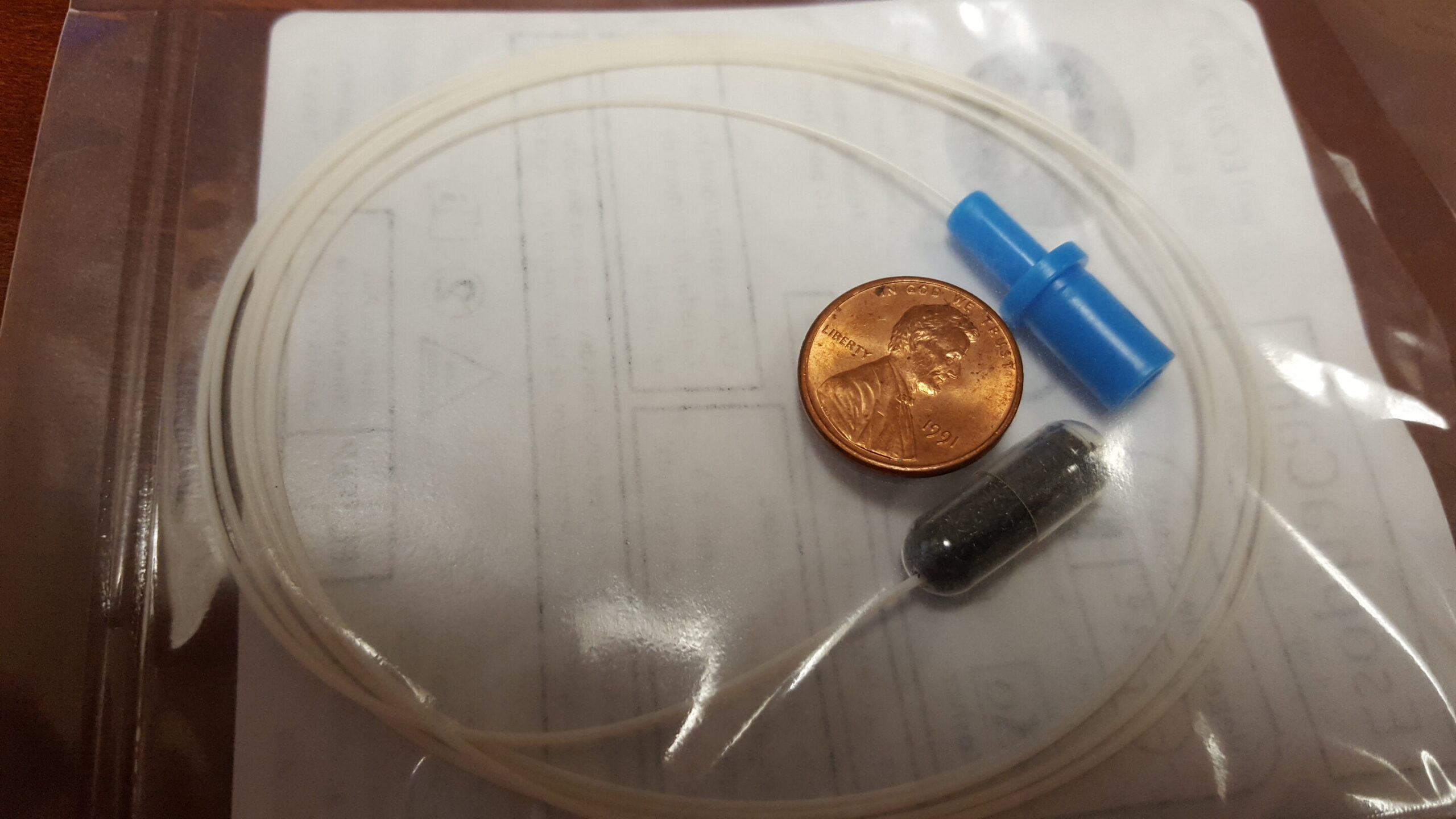
Researchers at the Johns Hopkins Kimmel Cancer Center and the Johns Hopkins University School of Medicine have developed a biomarker algorithm designed to identify esophageal cancer and precancerous conditions, such as Barrett’s esophagus (BE) and high-grade dysplasia. This innovative approach utilizes a noninvasive cell collection method, where patients swallow a capsule containing a compressed sponge attached to a string. Once the capsule reaches the stomach, the outer layer dissolves, allowing the sponge to expand. As the sponge is gently withdrawn, it collects cells from the esophagus, which are then analyzed for specific biomarker methylation patterns associated with esophageal malignancies.
The biomarker algorithm focuses on the methylation status of genes such as USP44, TBC1D30, and NELL1, which have been implicated in various cancers. For instance, USP44 has been identified as a marker in prostate, liver, and colorectal cancers, while TBC1D30 shows high methylation levels in colorectal cancer. By detecting these epigenetic modifications, the algorithm can differentiate between healthy tissue, BE, and esophageal adenocarcinoma. If validated through further studies, this method could serve as a less invasive screening tool, potentially identifying patients who would benefit from confirmatory endoscopic evaluations, thereby facilitating earlier detection and treatment of esophageal cancer. Click for More Details